A look into our
The Big Picture
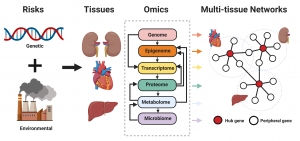
Common cardiometabolic disorders such as coronary artery disease (CAD), type 2 diabetes (T2D), non-alcoholic fatty liver disease (NAFLD), and obesity, and brain disorders such as Alzheimer’s disease (AD) and traumatic brain injury (TBI) represent top health concerns worldwide. These diseases are highly interconnected. We hypothesize that the complex interactions between genetic and environmental risk factors perturb specific gene networks which in turn induce variations in disease susceptibility and therapeutic response. Research in our lab employs integrative multiomics and systems biology approaches that leverage genetic, transcriptomic, epigenomic, proteomic, microbiomic, and phenotypic data from human and rodent populations to identify causal molecular networks that contribute to the development of complex diseases. We are also exploring the connection between cardiometabolic disorders with neuroplasticity and brain disorders. The candidate causal components and networks identified will serve as the basis for therapeutic target and biomarker discovery as well as for patient stratification with respect to clinical outcome or pharmacological response.
Hypothesis
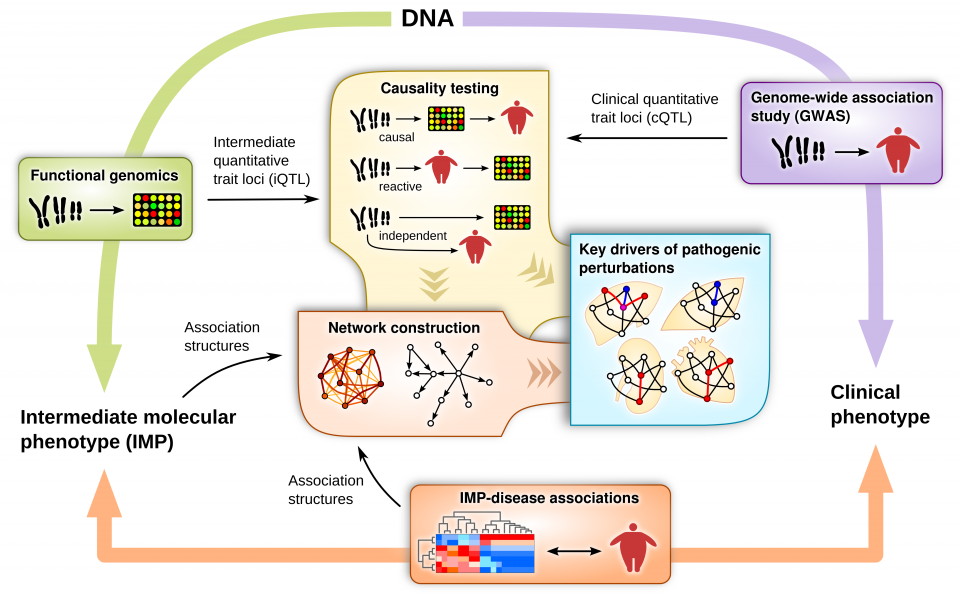
We hypothesize that genes, rather than acting in isolation, are organized into well-structured scale-free networks, in which central network genes or hubs play a more important role whereas peripheral genes play moderate to subtle roles. Common complex diseases/phenotypes conferred by genetic variations and environmental perturbations are mediated through specific parts of gene networks, termed subnetworks. Perturbation of a particular subnetwork may result in a specific disease or disease subtype and can be treated with a drug targeting the same subnetwork.
Figure credit: Montgomery Blencowe. Citation: Yang X. Multi-tissue Multi-omics Systems Biology of Complex Diseases. Trends in Molecular Medicine, 26:718-728, 2020. [link]
Approaches
Multiple layers of tissue-specific genomic and epigenomic information such as DNA variants, mRNA or protein expression, DNA methylation, gut microbiome, and metabolites, as well as phenotypic data are first collected from human or mouse populations in which variations in disease phenotypes are demonstrated. We then apply multiple integrative multiomics approaches such as causality testing, network modeling, and functional genomics to sort out the relationships between DNA variations, gene expression/epigenomic/protein level/gut bacteria community/metabolite changes, and phenotypic variations. Based on these relationships, we attempt to map the complex regulatory network structure underlying complex diseases.
Ongoing research directions
1. Identification of genetically-driven gene networks underlying individual diseases and across diseases.
This is achieved by comprehensive integration of genetic, gene expression, epigenetic, metabolite, and proteomic profiles of cells, tissues, and compound treatments relevant to each disease in animal models and human populations.
Representative publications:
Mäkinen V, Civelek M, Meng Q, Zhang B, Zhu J, Levian C,…Yang X*, Assimes T*. Integrative Genomics Reveals Novel Molecular Pathways and Gene Networks for Coronary Artery Disease. PLoS Genetics. 10(7):e1004502, 2014. [link] (Highlighed by Nature Reviews Genetics in “Piecing together the puzzle of cronary artery disease“).
Shu L, Chen KHK, Zhang G, Huan T, Kurt Z, Zhao Y, Codoni V, Tregouet DA, Yang J, Wilson JG, Luo X, Levy D, Lusis AJ, Liu S, Yang X. Shared genetic regulatory networks for cardiovascular disease and type 2 diabetes in multiple populations of diverse ethnicities in the United States. PLOS Genetics 3(9): e1007040, 2017. [link].
Chella Krishnan K+, Kurt Z+, Barrere-Cain R, Sabir S, Das A, Floyd R, Vergnes L, Zhao Y, Che N, Charugundla S, Qi H, Zhou Z, Meng Y, Pan C, Seldin MM, Norheim F, Hui S, Reue K, Lusis AJ, Yang X. Integration of Multi-omics Data from Mouse Diversity Panel Highlights Mitochondrial Dysfunction in Non-Alcoholic Fatty Liver Disease. Cell Systems 6:1-13, 2018. [link]
Kurt Z, Barrere-Cain R, LaGuardia J, Mehrabian M, Pan C, Hui ST, Norheim F, Zhou Z, Hasin Y, Lusis AJ, Yang X. Tissue-specific Pathways and Networks Underlying Sexual Dimorphism in Non-Alcoholic Fatty Liver Disease. Biology of Sex Differences, 9:46, 2018. [link]
Blencowe M+, Ahn IS+, Saleem Z, Luk H, Cely I, Makinen VP, Zhao Y*, Yang X*. Gene networks and pathways for plasma lipid traits via multi-tissue multi-omics systems analysis. Journal of Lipid Research, 62: 100019, 2021. [link]
2. Identification of environmentally-driven gene networks underlying complex diseases.
This is achieve by examining how environmental factors (eg., nutritional, obesogens) modulate the epigenome, transcriptome, and microbiome, and how they interact with genetic factors to affect diseases.
Representative publications:
, , , , , , , , , Prenatal Bisphenol A Exposure in Mice Induces Multi-tissue Multi-omics Disruptions Linking to Cardiometabolic Disorders. Endocrinology 160: 409-429, 2019. [link]
Diamante G, Cely I, Zamora Z, Ding J, Blencowe M, Lang J, Bline A, Singh M, Lusis AJ, Yang X. Systems Toxicogenomics of Prenatal Low-dose BPA Exposure on Liver Metabolic Pathways, Gut Microbiota, and Metabolic Health in Mice. Environment International 146: 106260, 2020. [link]
Zhang G, Meng Q, Blencowe M, Agrawal R, Gomez-Pinilla F, Yang X. Multi-tissue Multi-omics Nutrigenomics Indicates Context Specific Effects of DHA on Rat Brain. Molecular Nutrition and Food Research 64(23):2000788, 2020. [link]
Zuo Y, Iemolo A, Montilla-Perez A, Li H, Yang X, Telese F. Chronic adolescent exposure to cannabis in mice leads to long-term sex-biased changes in gene expression networks across brain regions. Neuropsychopharmacology, 2022. [link]
Ha S+, Ahn IS+, , ,,, , , Cross-tissue multiomics studies reveal gut-brain interactions mediating the effect of Akkermansia muciniphila in counteracting fructose-induced obesity. [bioRxiv link]
3. Investigation of the molecular networks that connect various diseases.
This is achieved by systems level integration of genetic and environmental risks that affect shared pathways and networks between diseases.
Representative publications:
Meng Q, Ying Z, Noble E, Zhao Y, Agrawal R, Mikhail A, Zhuang Y, Tyagi E, Zhang Q, Lee JH, Morselli M, Orozco L, Guo Q, Kilts TM, Zhu J, Zhang B, Pellegrini M, Xiao X, Young MF, Gomez-Pinilla F, Yang X. Systems Nutrigenomics Reveals Brain Gene Networks Linking Metabolic and Brain Disorders. EBioMedicine. 7:157-166, 2016. [link].
Meng Q, Zhuang Y, Ying Z, Agrawal R, Yang X*, Gomez-Pinilla F*. Traumatic Brain Injury Induces Genome-wide Transcriptomic, Methylomic, and Network Perturbations in Brain and Blood Predicting Neurological Disorders. EBioMedicine 16:184-194, 2017. [link].
, , , ,, , , Single Cell Molecular Alterations Reveal Target Cells and Pathways of Concussive Brain Injury. Nature Communications, 9:3894, 2018 [link]
Zuo Y, Wei D, Zhu C, Naveed O, Hong W, Yang X. Unveiling the pathogenesis of psychiatric disorders using network models. Genes, 12(7):1101, 2021. [link]
Arneson D+, Zhang G+, Ahn IS, Ying Z, Diamante G, Cely I, Palafox-Sanchez V, Gomez-Pinilla F, Yang X. Systems spatiotemporal dynamics of traumatic brain injury at single cell resolution reveals humanin as a therapeutic target. Cellular and Molecular Life Sciences, 79(9):480, 2022 [link]
4. Development of integrative genomics approaches for big data integration.
This is achieved by designing and implementing bioinformatics tools that effectively integrate multi-dimensional omics data (genetic, transcriptomic, epigenetic, metabolic, proteomic, etc) and model molecular networks and regulators of pathophysiology.
Representative publications:
Shu L, Zhao Y, Kurt Z, Byars SG, Tukiainen T, Kettunen J, Orozco, LD, Pellegrini M, Lusis AJ, Ripatti S, Zhang B, Inouye M, Makinen VP, Yang X. Mergeomics: Integration of Diverse Genomics Resources to Identify Pathogenic Perturbations to Biological Systems. BMC Genomics 17:874, 2016. [link to paper; link to R package at Mergeomics web server; link to R package at R Bioconductor].
Blencowe M, Arneson D, Ding J, Chen Y-W, Saleem Z, Yang X. Network Modeling of Single Cell Omics Data: Challenges, Opportunities, and Progress. Emerging Topics in Life Sciences 3 (4): 379-398, 2019. [link]
Littman R, Hemminger Z, Foreman R, Arneson D, Zhang G, Gomez-Pinilla F, Yang X*, Wollman R*. JSTA: joint cell segmentation and cell type annotation for spatial transcriptomics. Molecular Systems Biology 17:e10108, 2021. [link]
Ding J+, Blencowe M+, Nghiem TX+, Ha SM, Chen Y-W, Li G, Yang X. Mergeomics 2.0: A Web Server for Multi-omics Data Integration to Elucidate Disease Networks and Predict Therapeutics. Nucleic Acids Research 49: W375-387, 2021. [link]
Chen Y-W+, Diamante G+, Ding J+, Nghiem TX, Ha SM, Cohn P, Arneson D, Blencowe M, Garcia J, Zaghari N, Patel P, Yang J, Yang X. PharmOmics: A Species- and Tissue-specific Drug Signature Database and Online Tool for Drug Repurposing. iScience, 25(4):104052, 2022. [link]
Littman R, Wang N, Peng C, Yang X. SCING: Single Cell INtegrative Gene regulatory network inference elucidates robust, interpretable gene regulatory networks. [bioRxiv link]